CHARMM-GUI PACE CG Builder for Solution, Micelle, and Bilayer Coarse-Grained Simulations
By Yifei Qi, Xi Cheng, Wei Han, Sunhwan Jo, Klaus Schulten, and Wonpil Im.
Published in Journal of Chemical Information and Modeling 54(3) on March 4, 2014 (online publication date). PMID: 24624945. PMCID: PMCID3985889. Link to publication page.
Core Facility: Computational Modeling
Abstract
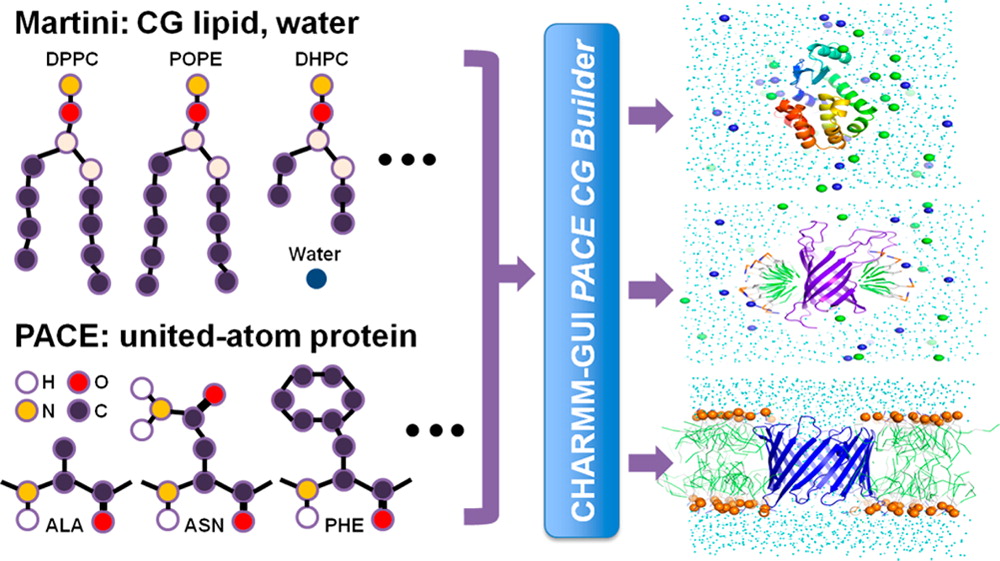
Coarse-grained (CG) and multiscale simulations are widely used to study large biological systems. However, preparing the simulation system is time-consuming when the system has multiple components, because each component must be arranged carefully as in protein/micelle or protein/bilayer systems. We have developed CHARMM-GUI PACE CG Builder for building solution, micelle, and bilayer systems using the PACE force field, a united-atom (UA) model for proteins, and the Martini CG force field for water, ions, and lipids. The robustness of PACE CG Builder is validated by simulations of various systems in solution (α3D, fibronectin, and lysozyme), micelles (Pf1, DAP12-NKG2C, OmpA, and DHPC-only micelle), and bilayers (GpA, OmpA, VDAC, MscL, OmpF, and lipid-only bilayers for six lipids). The micelle’s radius of gyration, the bilayer thickness, and the per-lipid area in bilayers are comparable to the values from previous all-atom and CG simulations. Most tested proteins have root-mean squared deviations of less than 3 Å. We expect PACE CG Builder to be a useful tool for modeling/refining large, complex biological systems at the mixed UA/CG level.




