Identification of a second substrate-binding site in solute-sodium symporters
By Zheng Li, Ashley S. E. Lee, Susanne Bracher, Heinrich Jung, Aviv Paz, Jay P. Kumar, Jeff Abramson, Matthias Quick, Lei Shi.
Published in Journal of Biological Chemistry, 2015 Jan 2;290(1):127-41. PMID: 25398883. PMCID: PMC4281715 [Available on 2016-01-02]. Link to publication page.
Projects: The Transport Cycle in Neurotransmitter Uptake Systems
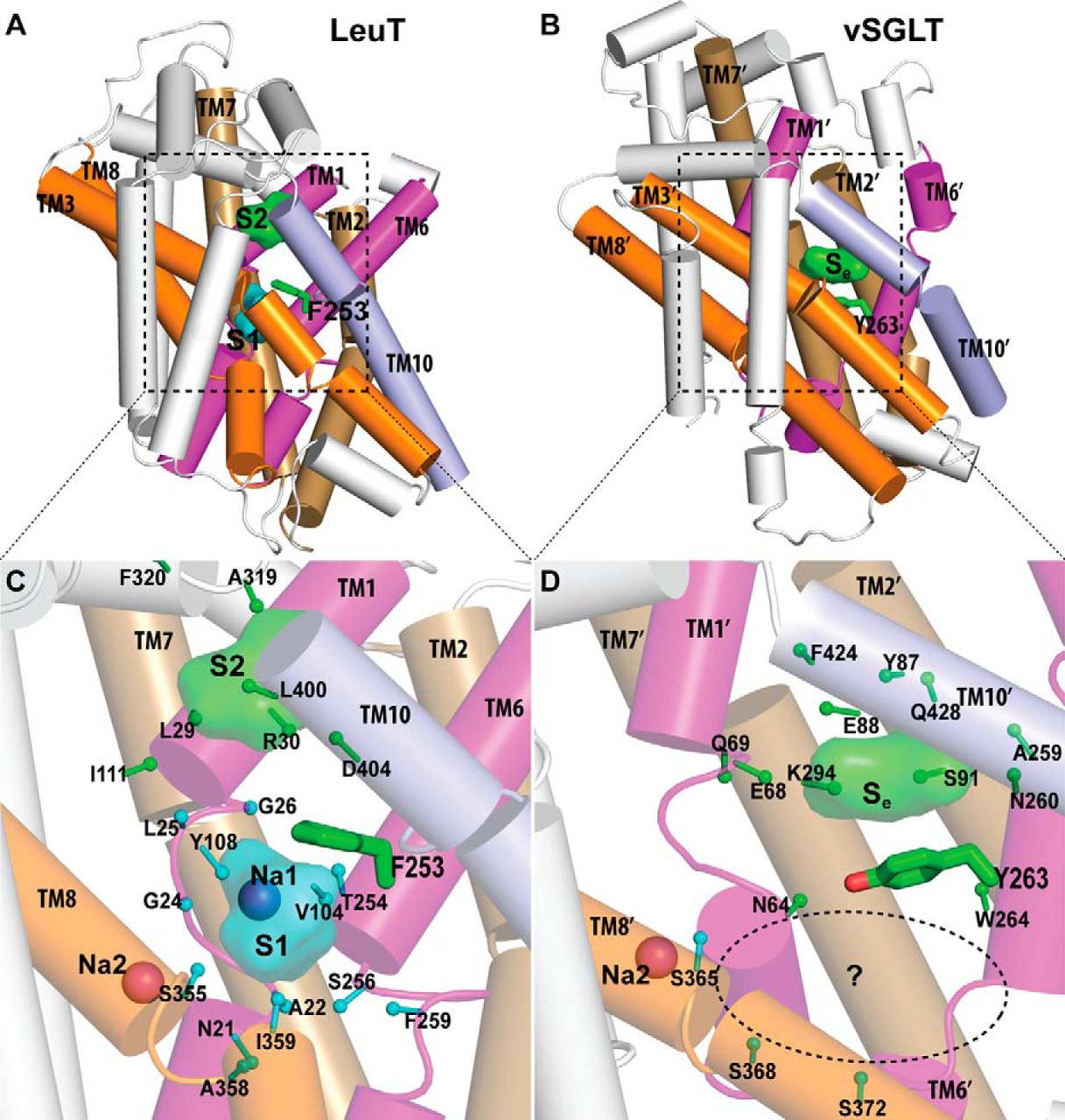
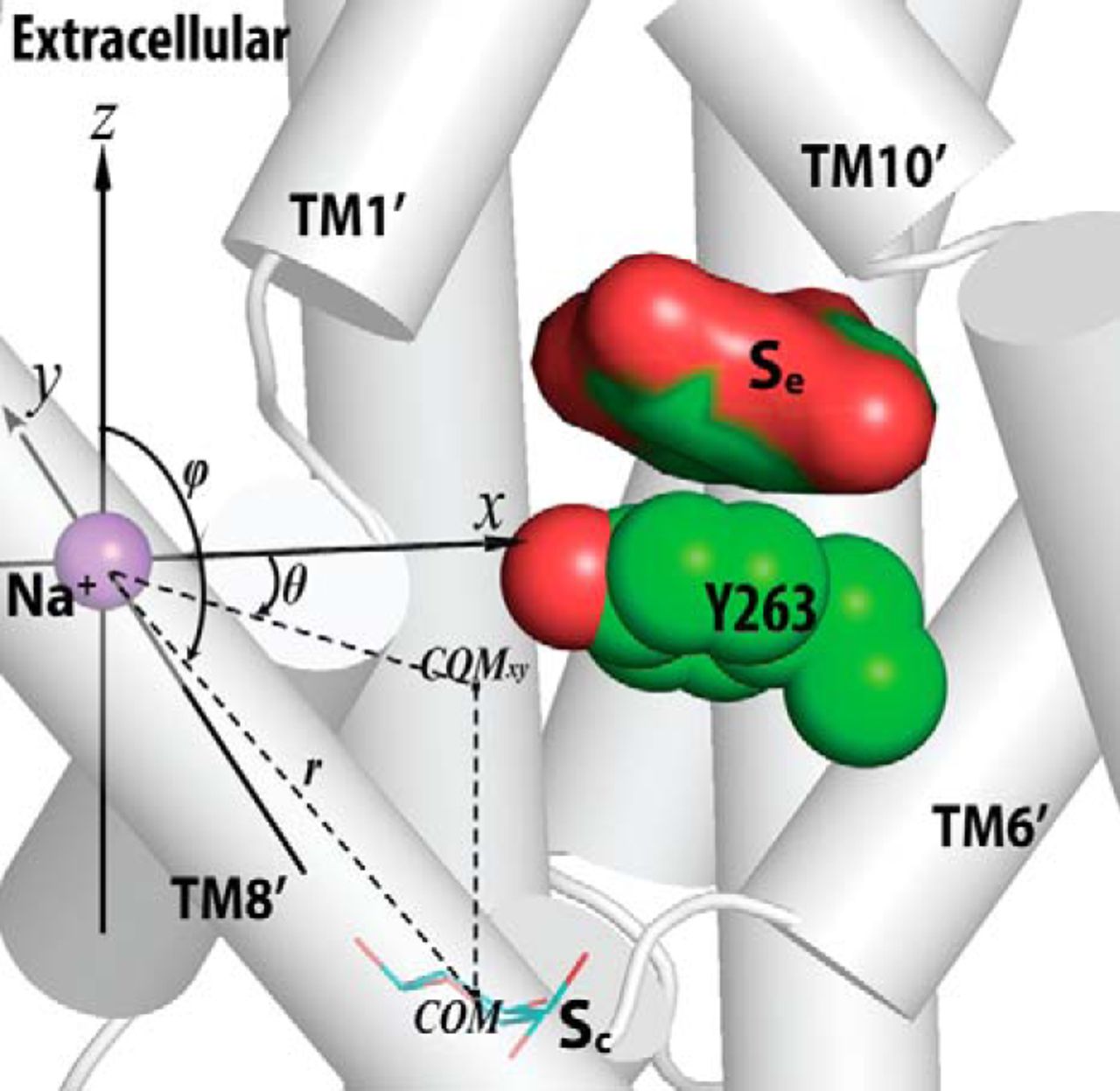
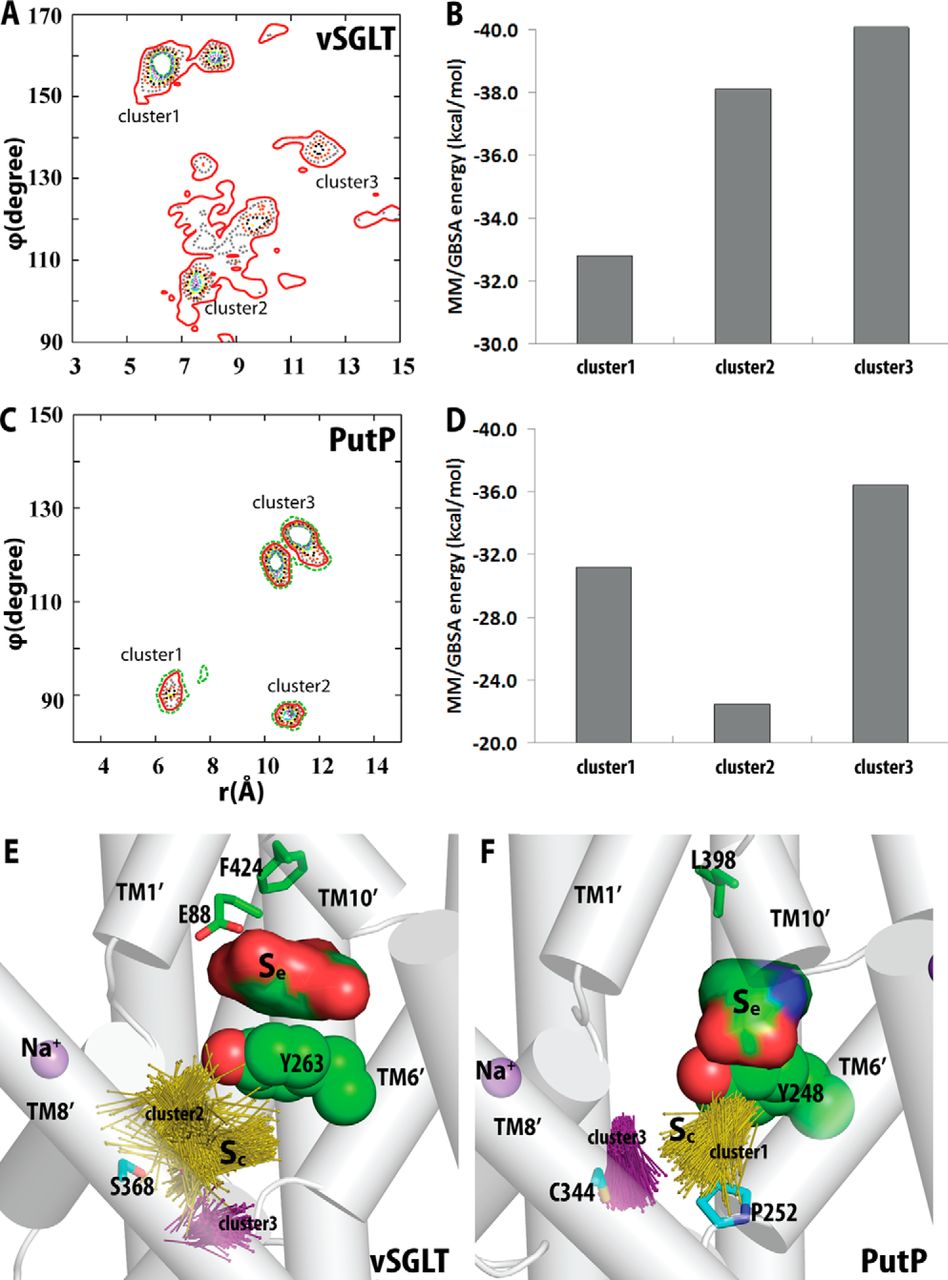
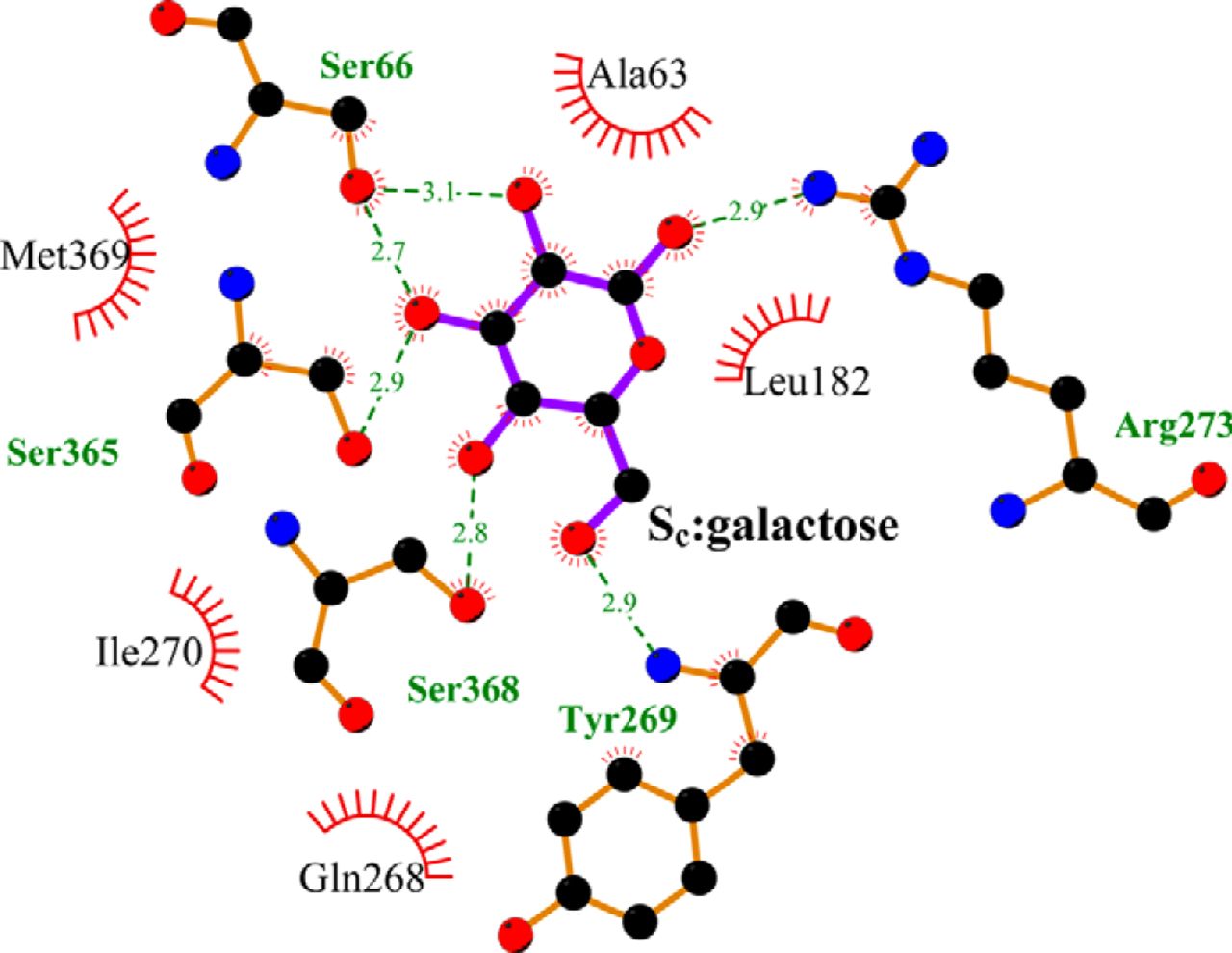
Figure 2. Crystallographically identified substrate-binding site of vSGLT is located more extracellularly than that of LeuT. A and B show the superimposed orientations of a LeuT model bound with two substrates Leu (24) and the vSGLT structure (PDB code 3DH4), respectively, based on a structure-structure alignment (see text). The TMs directly involved in substrate binding in either LeuT or vSGLT are in non-white colors and in the same color codes for both panels. In particular, the equivalent TMs in the two 5-TM inverted repeats of LeuT structural fold, e.g. TM1 and TM6, are in the same color. The substrate-binding site in the vSGLT structure, termed Se here, is enclosed by TM1′, -2′, -6′, -7′, and -10′, whereas for LeuT the crystallographically identified S1 site is formed by residues from TM1, -3, -6, and -8, and the computationally identified S2 site is formed by residues from TM 1, -3, -10, and EL4. C and D are the zoom-in views of the substrate-binding sites in LeuT and vSGLT, respectively. The cavity below Tyr-263 of vSGLT may potentially form a substrate-binding site. Thus Phe-253 of LeuT and Tyr-263 of vSGLT may play similar roles as the gating residues to separate more centrally located substrate-binding sites from the extracellular milieu. The Cα-Cβ bonds of substrate binding residues are shown as sticks (cyan for the S1/Sc site and green for the S2/Se sites) with the Cβ atoms represented as small spheres. D, residues Ser-365, Ser-368, and Ser-372 of vSGLT are aligned to Thr-341, Cys-344, and Val-348 of PutP, which have been found to be critical for proline uptake (42).
Abstract
The structure of the sodium/galactose transporter (vSGLT), a solute-sodium symporter (SSS) from Vibrio parahaemolyticus, shares a common structural fold with LeuT of the neurotransmitter-sodium symporter family. Structural alignments between LeuT and vSGLT reveal that the crystallographically identified galactose-binding site in vSGLT is located in a more extracellular location relative to the central substrate-binding site (S1) in LeuT. Our computational analyses suggest the existence of an additional galactose-binding site in vSGLT that aligns to the S1 site of LeuT. Radiolabeled galactose saturation binding experiments indicate that, like LeuT, vSGLT can simultaneously bind two substrate molecules under equilibrium conditions. Mutating key residues in the individual substrate-binding sites reduced the molar substrate-to-protein binding stoichiometry to ∼1. In addition, the related and more experimentally tractable SSS member PutP (the Na+/proline transporter) also exhibits a binding stoichiometry of 2. Targeting residues in the proposed sites with mutations results in the reduction of the binding stoichiometry and is accompanied by severely impaired translocation of proline. Our data suggest that substrate transport by SSS members requires both substrate-binding sites, thereby implying that SSSs and neurotransmitter-sodium symporters share common mechanistic elements in substrate transport.