Computational characterization of structural dynamics underlying function in active membrane transporters
By Jing Li, Po-Chao Wen, Mahmoud Moradi, and Emad Tajkhorshid.
Published in Curr Opin Struct Biol. 2015 Apr;31:96-105. Epub 2015 Apr 27.
PMID: 25913536. PMCID: PMC4476910 [Available on 2016-04-27]. Link to publication page.
Core: Computational Modeling Core.
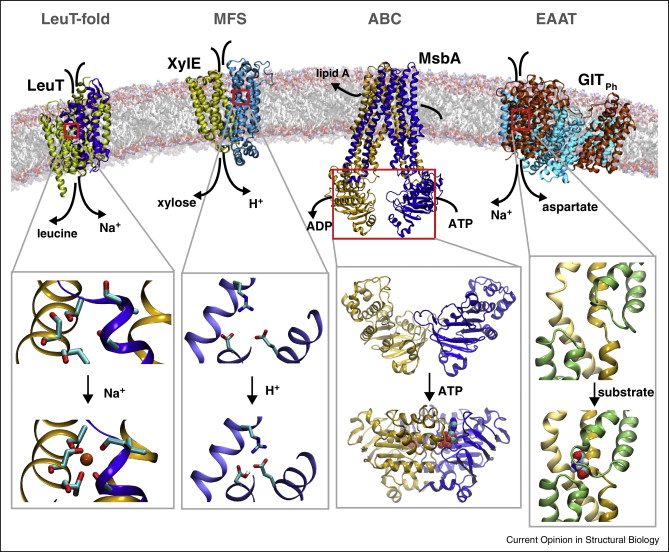
Abstract
Active transport of materials across the cellular membrane is one the most fundamental processes in biology. In order to accomplish this task, membrane transporters rely on a wide range of conformational changes spanning multiple time and size scales. These molecular events govern key functional aspects in membrane transporters, namely, coordinated gating motions underlying the alternating access mode of operation, and coupling of uphill transport of substrate to various sources of energy, for example, transmembrane electrochemical gradients and ATP binding and hydrolysis. Computational techniques such as molecular dynamics simulations and free energy calculations have equipped us with a powerful repertoire of biophysical tools offering unparalleled spatial and temporal resolutions that can effectively complement experimental methodologies, and therefore help fill the gap of knowledge in understanding the molecular basis of function in membrane transporters.

![Figure 2. Monitoring water dynamics within proteins brings novel insights into transport mechanism. (Left) The continuous water wire spanning the ClC<sub>ec1</sub> lumen depend on the presence of Cl<sup>−</sup> (green sphere), as the H+ transport pathway. See [37•] for further details. (Right) Transient water-conducting state observed in vSGLT, which is permeable to water but occluded to the substrate (orange van der Waals). See [39••] for further details.](/site-media/images/publications/2015/li_wen2015-figure2.jpg)
![Figure 3. Example of protein-lipid interactions captured in MD simulations. (a) The protrusion of an annular lipid into the substrate binding site of Pgp, which is a known lipid flippase. (b) Close-up view of the protrusion site along the crevice opening. See [22] for further details.](/site-media/images/publications/2015/li_wen2015-figure3.jpg)
![Figure 4. Investigating the OF→IF transition in MsbA transporter using system-specific reaction coordinates α, β, and γ, describing protein global conformation [58••]. (a) Multiple nonequilibrium MD trajectories generated using different biasing protocols and projected onto the reduced space of (α, β, γ). (b) Nonequilibrium work required to induce the OF→IF transition associated with trajectories shown in (a). (c) Snapshots of MsbA structure along the optimum pathway shown in (b). (d) The water density along the pore measured for the optimum pathway shown in (c). These results are in line with alternating access mechanism.](/site-media/images/publications/2015/li_wen2015-figure4.jpg)

