Conformational Chaperones for Structural Studies of Membrane Proteins Using Antibody Phage Display with Nanodiscs
By Pawel K. Dominik, Marta T. Borowska, Olivier Dalmas, Sangwoo S. Kim, Eduardo Perozo, Robert J. Keenan, and Anthony A. Kossiakoff.
Published in Structure. 2015 Dec 28. pii: S0969-2126(15)00503-1. [Epub ahead of print]. PMID: 26749445. PMCID: 4740257. Link to Pubmed page.
Core Facility: Synthetic Antigen Binder (SAB) Generation and Crystallography and Protein Production Core.
Abstract
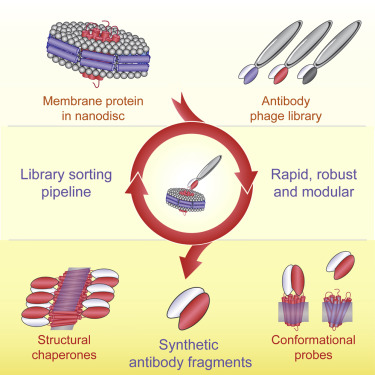
A major challenge in membrane biophysics is to define the mechanistic linkages between a protein’s conformational transitions and its function. We describe a novel approach to stabilize transient functional states of membrane proteins in native-like lipid environments allowing for their structural and biochemical characterization. This is accomplished by combining the power of antibody Fab-based phage display selection with the benefits of embedding membrane protein targets in lipid-filled nanodiscs. In addition to providing a stabilizing lipid environment, nanodiscs afford significant technical advantages over detergent-based formats. This enables the production of a rich pool of high-performance Fab binders that can be used as crystallization chaperones, as fiducial markers for single-particle cryoelectron microscopy, and as probes of different conformational states. Moreover, nanodisc-generated Fabs can be used to identify detergents that best mimic native membrane environments for use in biophysical studies.
Highlights
- Nanodiscs allow for generation of multiple antibody fragments to membrane proteins
- Improved phage display protocol is fast, efficient, and modular
- Resulting antibodies probe protein conformations present in lipid environment
- Expands the toolbox for structural and functional studies of membrane proteins


