Multi-ion free energy landscapes underscore the microscopic mechanism of ion selectivity in the KcsA channel
By David Medovoy, Eduardo Perozo, Benoît Roux.
Published in Biochim Biophys Acta 2016 Jul;1858(7 Pt B):1722-32. PMID: 26896693. PMCID: PMC4939264. Link to Pubmed page.
Project: Dynamics of Ion Permeation and Conformational Coupling in – KcsA. Core Facility: Computational Modeling.
Abstract
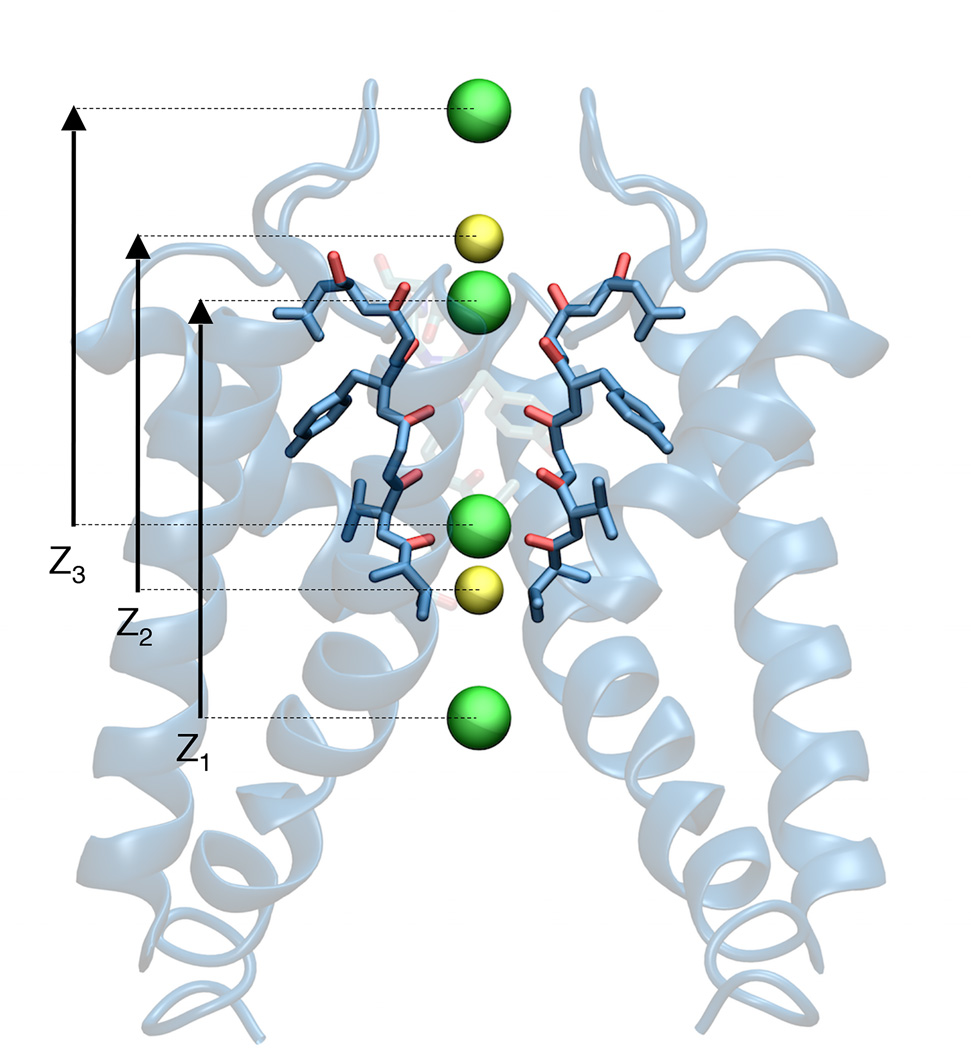
Potassium (K(+)) channels are transmembrane proteins that passively and selectively allow K(+) ions to flow through them, after opening in response to an external stimulus. One of the most critical functional aspects of their function is their ability to remain very selective for K(+) over Na(+) while allowing high-throughput ion conduction at a rate close to the diffusion limit. Classically, it is assumed that the free energy difference between K(+) and Na(+) in the pore relative to the bulk solution is the critical quantity at the origin of selectivity. This is the thermodynamic view of ion selectivity. An alternative view assumes that kinetic factors play the dominant role. Recent results from a number of studies have also highlighted the great importance of the multi-ion single file on the selectivity of K(+) channels. The data indicate that having multiple K(+) ions bound simultaneously is required for selective K(+) conduction, and that a reduction in the number of bound K(+) ions destroys the multi-ion selectivity mechanism utilized by K(+) channels. In the present study, multi-ion potential of mean force molecular dynamics computations are carried out to clarify the mechanism of ion selectivity in the KcsA channel. The computations show that the multi-ion character of the permeation process is a critical element for establishing the selective ion conductivity through K(+)-channels. This article is part of a Special Issue entitled: Membrane Proteins edited by J.C. Gumbart and Sergei Noskov.


