MPSDC to host Frontiers in Membrane Protein Dynamics 2014
Continuing with our successful annual meetings and workshops over the past three years, the Membrane Protein Structural Dynamics Consortium (MPSDC) announces the second Frontiers in Membrane Protein Dynamics meeting in 2014. The meeting, which will take place in the Chicago Hilton Hotel in the downtown Chicago loop area, is open to the public and will be our most extensive meeting to date.
Like our first Frontiers in Membrane Protein Dynamics meeting in 2012, the conference will consist of scientific sessions and poster presentations, and feature both Consortium members and external invitees. Unlike in previous years, however, Frontiers in Membrane Protein Dynamics 2014 will span three days of scientific discussion, organized into eight sessions covering topics at the edge of current developments in membrane protein structure, dynamics and function.
 Additionally, we are thrilled to announce that Frontiers in Membrane Protein Dynamics 2014 will feature two keynote speakers. On May 7th, the meeting will be inaugurated by Dr. Robert Stroud, Professor of Biochemistry & Biophysics and Pharmaceutical Chemistry at the University of California at San Francisco. Additionally, we are thrilled to announce that Frontiers in Membrane Protein Dynamics 2014 will feature two keynote speakers. On May 7th, the meeting will be inaugurated by Dr. Robert Stroud, Professor of Biochemistry & Biophysics and Pharmaceutical Chemistry at the University of California at San Francisco.
 Then, on May 9th, Dr. Klaus Schulten, Swanlund Professor of Physics at the University of Illinois at Urbana Champaign and an active participant in the Consortium’s own Computational Modeling Core, will close the conference. Both Stroud and Schulten are renowned experts at the vanguard of membrane protein dynamics and their unique insights on the contemporary state of the scientific field will significantly enhance our discussions. Read more about Drs. Stroud and Schulten » Then, on May 9th, Dr. Klaus Schulten, Swanlund Professor of Physics at the University of Illinois at Urbana Champaign and an active participant in the Consortium’s own Computational Modeling Core, will close the conference. Both Stroud and Schulten are renowned experts at the vanguard of membrane protein dynamics and their unique insights on the contemporary state of the scientific field will significantly enhance our discussions. Read more about Drs. Stroud and Schulten »
Finally, we count a stellar roster of speakers covering a broad range of topics and techniques. A preliminary speaker list can be found here »
Up-to-date information about Frontiers in Membrane Protein Dynamics 2014, can be found at a stand-alone website portal dedicated exclusively to this meeting. We invite you to visit the Frontiers in Membrane Protein Dynamics 2014 website, and let us know what you think! Please stay posted as we will be releasing more information to the website soon.
 Visit the website » Visit the website »
Back to top 
MPSDC Protein Expression and Purification core adds two scientists to the team
We are pleased to announce that Drs. Francis Valiyaveetil and Andrzej Rajca and their research teams have officially joined the Consortium. Valiyaveetil and Rajca began their collaborations with the MPSDC as Associate Members, and are now participating as active members of the Membrane Protein Expression/Purification Core.
 Francis Valiyaveetil is Assistant Professor of Physiology & Pharmacology at Oregon Health & Science University. The Valiyaveetil lab studies potassium channels, which are integral membrane proteins that catalyze the selective conduction of K+ ions across biological membranes. While a great deal of research has been focused on these channels, fundamental questions regarding the mechanism of ionic selectivity and gating remain. Valiyaveetil’s team has developed a unique combination of methods to address these questions. Their methods include the use of chemical synthesis to introduce precise chemical changes in the channels, x-ray crystallography to determine the structural effects and electrophysiology to determine the functional effects of these changes. Using this multidisciplinary approach they hope to explain the mechanism of ion selectivity and channel gating. Francis Valiyaveetil is Assistant Professor of Physiology & Pharmacology at Oregon Health & Science University. The Valiyaveetil lab studies potassium channels, which are integral membrane proteins that catalyze the selective conduction of K+ ions across biological membranes. While a great deal of research has been focused on these channels, fundamental questions regarding the mechanism of ionic selectivity and gating remain. Valiyaveetil’s team has developed a unique combination of methods to address these questions. Their methods include the use of chemical synthesis to introduce precise chemical changes in the channels, x-ray crystallography to determine the structural effects and electrophysiology to determine the functional effects of these changes. Using this multidisciplinary approach they hope to explain the mechanism of ion selectivity and channel gating.
 Andrzej Rajca is Charles Bessey Professor of Chemistry at University of Nebraska. The Rajca laboratory research interests lie in the area of organic and biomaterials chemistry. The underlying theme of the laboratory’s research is rational design and synthesis of new materials (molecules and polymers) with magnetic and optical properties. Rational design of new structures and materials (molecules and polymers) with targeted properties are implemented using the tools of organic synthesis, crystallography, magnetic resonance spectroscopy (NMR and EPR) and imaging (MRI), as well as magnetometry (SQUID), circular dichroism (CD) spectroscopy and multiangle light scattering (MALS). Andrzej Rajca is Charles Bessey Professor of Chemistry at University of Nebraska. The Rajca laboratory research interests lie in the area of organic and biomaterials chemistry. The underlying theme of the laboratory’s research is rational design and synthesis of new materials (molecules and polymers) with magnetic and optical properties. Rational design of new structures and materials (molecules and polymers) with targeted properties are implemented using the tools of organic synthesis, crystallography, magnetic resonance spectroscopy (NMR and EPR) and imaging (MRI), as well as magnetometry (SQUID), circular dichroism (CD) spectroscopy and multiangle light scattering (MALS).
According to Robert Nakamoto, PI of the Protein core, Rajca and Valiyaveetil will play a pivotal role in the core’s ongoing research activities: “Andrzej Rajca provides expertise in organic synthesis of nitroxide-containing compounds, and Frances Valiyaveetil is developing in vitro methods for synthesis and folding of membranous polypeptides. These investigators are collaborating with Protein Core members to develop methods for incorporation of spectroscopic labels directly into target proteins.”
Welcome, Andrzej and Francis!
Back to top

Consortium progress and scientific advances discussed at 2013 annual meeting

For the third year in a row, the MPSDC hosted a number of events in its home base of Chicago during the month of May, key among these its third annual meeting. Unlike last year’s Frontiers in Membrane Protein Structural Dynamics conference, this year’s annual meeting was closed to the public, although there are plans to host a second open attendance conference in the near future.
The MPSDC’s annual meeting allows for Consortium PI’s to present on the latest advances in the respective cores and projects, and serves as a dedicated time and space for members to discuss their research and organically find ways to collaborate with colleagues. Moreover, it gives the executive arm of the MPSDC the opportunity to report on “the state of the Consortium.”
At this year’s annual meeting at the University of Chicago’s Gleacher Center, MPSDC director Eduardo Perozo highlighted the continual exchange of ideas within and dynamics of the Consortium itself. Focusing specifically on the activities of the past year’s accomplishments, Dr. Perozo discussed the rational and efficient consolidation of Core Facilities to their highest efficiency and optimal productivity in close collaboration with individual projects. Followed by Dr. Perozo’s framing discussion, the PI’s of each of the three cores and seven projects described the progress made and latest scientific findings in their respective teams, providing attendees with the opportunity to respond and provide helpful feedback. Additionally, we invited several of our associate members to present talks, in order to foster further cross-pollination between our research and theirs.
In sum, we are thrilled to report that the Consortium is thriving, from both a strictly scientific stand point as well as in regards to our output to the community. We’d like to thank all who attended and partook in this year’s discussions, and look forward to seeing you next year!
 Read more » Read more »
Back to top 
Core workshops and minisymposium highlight ongoing research
As in previous years, the MPSDC's cores sponsored several satellite events in Chicago area prior to the Consortium's annual meeting. This year, the Computational Modeling Core hosted a membrane protein modeling workshop, and a mini-symposium concerning the latest advances in computational approaches to the study of membrane proteins. As before, the modeling workshop provided attendants with an overview of the use of the modeling dynamics and visualization software NAMD and VMD, and also featured Dr. Wonpil Im’s CHARMM-GUI Ligand Binder module. This year’s mini-symposium covered a number of topics including force field and atomic models, structural modeling with low-resolution data, and transition pathways. The minisymposium hosted a “keynote” lecture of sorts on 2D-IR Spectroscopy, held by Josh Carr from the University of Wisconson-Madison. We are pleased to present you with a recording of Carr’s lecture, titled Connecting Experiment and Simulation by Modeling the Protein Amide I Band.
 Watch the video » Watch the video »
This year, the Consortium’s Membrane Protein Expression/Purification Core held its first workshop as well. This workshop featured several Consortium collaborators such as Edith Buchinger from Goethe University, Stephen Pless from the University of British Columbia, Lilie Leisle from the University of Iowa, and Andrzej Rajca from the University of Nebraska. Topics discussed at this workshop included cellular and cell-free production of membrane proteins, reconstitution, incorporation of unnatural amino acids, single antigen binder technologies, and chemistry of protein modification and nitroxide spin labels. Both workshops and minisymposium were well attended and productive, and we will continue to host such satellite events in the future.
Back to top

Transporter linked to autism risk
MPSDC team member Hassane Mchaourab was recently featured in the Vanderbilt University Medical Center Reporter, along with a team of scientists who have linked a non-inherited, de novo mutation in the dopamine transporter to autism spectrum disorder (ASD).
The research was partially funded by the MPSDC, and contributes to the overall objectives of the Transport Cycle in Neurotransmitter Uptake Systems project in which Mchaourab is an active collaborator.
The group's research was published in the journal Molecular Psychiatry, with Mchaourab as one of the senior authors. You can read more about the publication here.
 Heiner Matthies, Ph.D., at the white board, leads a “seminar” for colleagues who hold vials of their fruit fly model that for the first time linked a non-inherited mutation in the dopamine transporter to autism. Seated at right, from the back, are Nicholas Campbell, Aurelio Galli, Ph.D., and P.J. Hamilton. Seated at left are Hassane Mchaourab, Ph.D., and James Sutcliffe, Ph.D. (photo by Susan Urmy)
 Read more » Read more »
Back to top

Collaborative MPSDC team develops innovative computational simulation technique
 We are delighted to share with you the news about a new computational simulation technique developed by several MPSDC team members that was first presented at last year’s Frontiers in Membrane Protein Dynamics conference. The development of this technique speaks to the significant scientific collaborations that take place under the umbrella of the Consortium, as well as the scientific conversations that began in Chicago last year. We are delighted to share with you the news about a new computational simulation technique developed by several MPSDC team members that was first presented at last year’s Frontiers in Membrane Protein Dynamics conference. The development of this technique speaks to the significant scientific collaborations that take place under the umbrella of the Consortium, as well as the scientific conversations that began in Chicago last year.
At the conference, Benoît Roux from our Computational Modeling Core introduced his team’s findings obtained from DEER (Double Electron-Electronic Resonance) data. At the conference, Roux and his team received helpful feedback from a number of scholars affiliated with the MPSDC as well as external invitees. After the conference, Roux and his team collaborated with a number of other scientists, including consortium colleague Hassane Mchaourab, to develop a novel computational simulation technique for exploiting the information from distance distribution data obtained from ESR/DEER spectroscopy for the refinement of membrane protein structures.
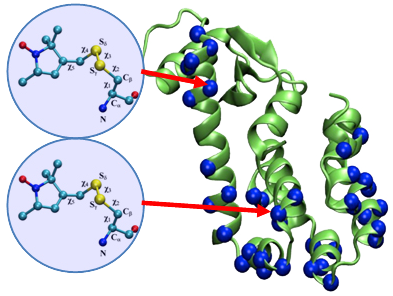 This simulation technique, called the Restrained-Ensemble Molecular Dynamics (REMD) simulation method, uses a global ensemble-based energy restraint to force the spin-spin distance distribution histograms calculated from a multiple-copy molecular dynamics simulation to match those obtained from ESR/DEER experiments.
Already, the method has yielded three unique publications detailing the results of these experiments, one of which (co-authored by Roux and Dr. Jean Weare) was highlighted by the Journal of Chemical Physics on their Top 20 Most Read in March 2013. Roux and his team have also gone on to apply this method to VSD (voltage-sensing domain) data with Eduardo Perozo, and Glt(Ph) data with Olga Boudker. Additionally, Wonpil Im is also implementing an easy setup of this method with dummy spin-labels on his CHARMM-GUI generator.
Shahidul M. Islam from Roux’s team, who co-authored two of the above papers and has been deeply involved in the scientific process, provided the MPSDC with an overview of the technique and its utility. We invite you to read his overview here »
Congratulations to all involved in the development of this exciting and important new method!
Back to top

Video feature: Behind the scenes in Emad Tajkhorshid's laboratory
Now available: our latest episode of our Behind the Scenes series. Within this series, we explore the inner workings of the Consortium with a camera in hand, dedicating to showing you what takes place within the laboratory setting and outside of it.
For our second feature, we present the following video feature of Emad Tajkhorshid, an active participant in the Consortium's Computational Modeling Core and the Structural Dynamics of ABC Transporter and Conformational Dynamics in the CLC Channel/Transporter Family projects. We sat down with Emad and asked him about his work with computational modeling, how his laboratory collaborates with other entities within the Consortium, and how he views the role of the Consortium within the broader scientific community. Let us know what you think in the comments!

 Watch the video » Watch the video »
Back to top
|
